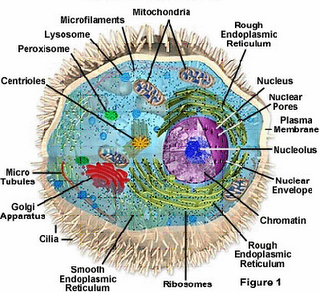
One of the fundamental goals in cell biology and proteomics is to identify the functions of proteins in the context of compartments that organize them in the cellular environment. Knowledge of subcellular locations of proteins can provide key hints for revealing their functions and understanding how they interact with each other in cellular networking. Unfortunately, it is both time-consuming and expensive to determine the localization of an uncharacterized protein in a living cell purely based on experiments.
Location classificationAccording to their subcellular locations, proteins are classified
into the following 12 discriminative groups: (1) chloroplast,(2) cytoplasm, (3) cytoskeleton, (4) endoplasmic reticulum, (5) extracell, (6) Golgi apparatus, (7) lysosome, (8) mitochondria,(9) nucleus, (10) peroxisome, (11) plasma membrane and (12) vacuole
Such a classification covers almost all the organelles in an animal or plant cell . With the rapid increase in new protein sequences entering into data banks, we are confronted with a challenge: is it possible to utilize a bioinformatic approach to help expedite the determination of protein subcellular locations?The enormous complexity of the protein sorting process, alternative means of transportation pathways, and lack of complete data for every organelle, present great challenges to the eager prediction method developers.
Categories of computational predictorsComputational methods for predicting protein sub-cellular localization can generally be divided into four categories: prediction methods based on
(i) The over all protein amino acid composition,
(ii) Known targeting sequences
(iii) Sequence homology and/or motifs,and
(iv) A combination of several sources of information from the first three categories
(hybrid methods).
Database of Protein subcellular localizationDBSubLoc:
http://www.bioinfo.tsinghua.edu.cn/~guotao/intro.htmlOnline Prediction toolsCello
http://cello.life.nctu.edu.tw/ChloroP
http://www.cbs.dtu.dk/services/ChloroP/ESLpred
http://www.imtech.res.in/raghava/eslpred/iPSORT
http://hc.ims.u-tokyo.ac.jp/iPSORT/MITOPRED
http://mitopred.sdsc.edu/predictNLS
http://cubic.bioc.columbia.edu/predictNLS/Proteome Analyst
http://www.cs.ualberta.ca/%7Ebioinfo/PA/Sub/index.htmlPSORT
http://psort.ims.u-tokyo.ac.jp/form.htmlPSORT II
http://psort.ims.u-tokyo.ac.jp/form2.htmlPSORT-B
http://www.psort.org/psortb/SignalP
http://www.cbs.dtu.dk/services/SignalP/SubLoc
http://www.bioinfo.tsinghua.edu.cn/SubLoc/TargetP
http://www.cbs.dtu.dk/services/TargetP/References:Dönnes, P, and Höglund, A (2004). Predicting Protein Subcellular Localization: Past, Present, and Future Genomics Proteomics Bioinformatics 2(4):209--215.
Kuo-Chen Chou1 and David W. Elrod (1999).Protein subcellular location prediction Protein Engineering 12(2): 107-118